Abstract
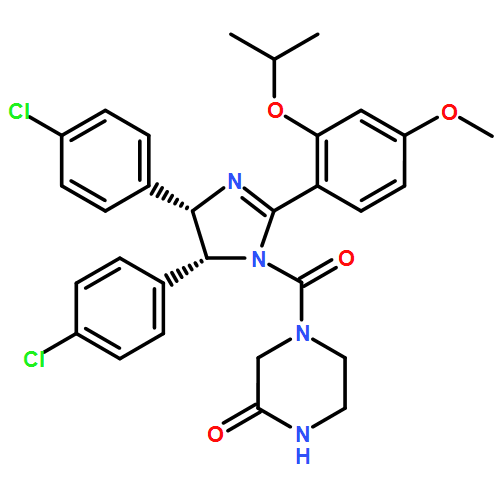
α-Helix-mediated protein–protein interactions (PPIs) are important targets for small-molecule inhibition; however, generic approaches to inhibitor design are in their infancy and would benefit from QSAR analyses to rationalise the noncovalent basis of molecular recognition by designed ligands. Using a helix mimetic based on an oligoamide scaffold, we have exploited the power of a modular synthesis to access compounds that can readily be used to understand the noncovalent determinants of hDM2 recognition by this series of cell-active p53/hDM2 inhibitors.